NIGMS Biomedical Technology Optimization and Dissemination (BTOD) Centers Program
DOROTHY BECKETT: OK. I think we'll get started. My name is Dorothy Beckett, and I'm the Director of the BBCB Division here at NIGMS. Welcome to today's webinar on the Biomedical Technology Optimization and Dissemination Centers. Our goal today is to provide you with information about these centers, both what our intent is for the program, how the program is structured. But also, we are very fortunate to have two PIs of active centers Vicki Wysocki and Ryan Mehl, who will tell you about how they are implementing that structure in their programs.
So next slide, please. So we have a group of presenters from NIGMS who will get things started. Of course, I just introduce myself, Dorothy Beckett. Christina Liu, Alvin Yeh, and Miljan Simonovic will be telling you all about the program and the technologies, and they will then say-- the webinar will then send you into the presentations from our two PIs. Next slide, please. So just a few housekeeping items.
I want to emphasize that today's webinar is informational. We'd like you to get a really clear idea of what the goals of the BTOD program are, and what is the structural framework that we have developed to achieve those goals, and then, as I said previously, how are these implemented in two active centers. The technologies as you'll see in the talk are really broad, and so the specifics of how the goals and structure are implemented are not necessarily the same for each center. And finally, if you have questions about application preparation, please submit them to the address shown on this slide. (NIGMS_BTODMailbox@nigms.nih.gov) We'll show you this again just to remind you. OK.
So to get things started in terms of the actual meat of the webinar, Christina Liu is going to kick it off. CHRISTINA LIU: Next slide, please. Thanks, Dorothy. The outline of today's webinar is here.
First, the program will first provide the overview of the BTOD program. There will be presentations from two active BTOD center PIs about their centers. And please note, all Q&A will be held at the end. So please enter your questions in the chat during our presentations. Next slide, please.
Before diving into the BTOD program description, let me describe the technology development pipeline, and how NIGMS supports this pipeline from its early stages of development to the broad dissemination of products to the research community. During the early stage of technology development, exploratory research for technology development, R21, supports proof of concept efforts. Once the proof of concept has been achieved, focused technology research development, R01, will support developing and validating prototype technology. After the prototype technology has been validated and its utility has been demonstrated, the RM1 BTOD center program will support prototype optimization and late-stage development toward broad dissemination.
Next slide, please. Today's webinar is about the BTOD center program, which aims to optimize state-of-the-art, late-stage technologies, and disseminate these technologies to be used by a broad range of biomedical researchers. Next slide, please.
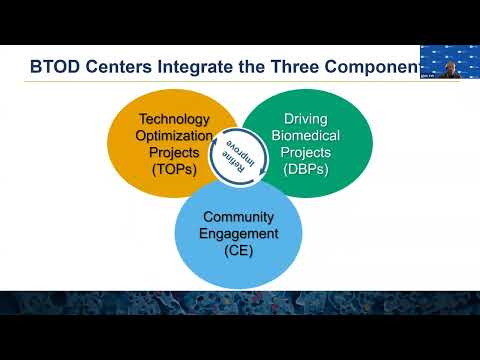
There are three components that are required to achieve the BTOD center program goals. The first component is technology optimization projects, we call it TOPs. And the second component is driving biomedical projects, called DBPs. And the third one is community engagement, we call CE. Alvin will take over to describe this individual components and how these components interact in the following slides. Alvin, take it away.
ALVIN YEH: Thanks, Christina. Next slide, please. So there may be up to three technology optimization projects within the center, and these are the central focus of the BTOD. The technologies should be later stage and within NIGMS mission. The projects may be multiple applications of a technology, or projects may be complementary technologies with a common goal.
The focus of the project should have potential for broad adoption by experts and nonexperts. Next slide. There may be up to 10 driving biomedical projects in the application, and these are independently funded, meaning that they've already been peer reviewed. And so their evaluation is really on how well they add to the optimization of the technologies. In that sense then, they should present challenging biomedical problems or technical environments, and they should be drawn from diverse nationally distributed research labs. These driving biomedical projects should be short-term collaborations with the center itself, and so it is expected that these projects turn over during the project period.
Next slide. So the ultimate goal of the center is to disseminate these optimized technologies to the broader research communities. So this component, the community engagement develops self-sustaining activities to disseminate the optimized technologies. These activities may include training, publications, seminars, workshops, and commercialization. We should emphasize that these activities should be self-sustaining because the center itself has a maximum lifetime of 15 years-- of up to 15 years with an initial project period of up to five years. Next slide.
And so the BTOD center then integrates these three components of technology, optimization, and collaborating with driving biomedical projects, and being disseminated through community engagement. And so it's really the scope of the technologies themselves that lead to the recruitment of both the projects and the broader research community, and it's really critical for the community engagement or the center to engage with the community to disseminate these technologies. Next slide.
I think I'm going to be turning it over to Miljan now. MILJAN SIMONOVIC: Yeah. Thanks. Thanks, Alvin, for providing the overview of components of BTOD center program. Now, many of you are probably wondering what are those features of your technology that may be attractive for this particular funding mechanism, and here we are just providing in this slide a snapshot.
What we are interested in is a technology that is in current state of the art, place, and that's distinct in its capabilities, and that you've demonstrated utility. That means that you publish some work where it's clear that that technology can be applied. Now, we don't want something that is really, really super, super mature. We want technologies that also require some optimization and fine tuning because there will be different biological questions, different platforms, different set of problems for different PIs.
We want to see that those technologies can be further refined, modified, and then applied to a variety of biomedical problems. And I emphasize the potential of that utility for a variety of problems. You don't want the technology that is so unique it's a niche technology. We want something that can be taken out from your lab and disseminate it out across the nation to different labs, potentially in a way of being portable so that other labs can take your technology in terms of a standardized protocol and apply to their particular question. And the last and very important thing that in NIGMS mission-- next slide, please.
So let me remind you what the NIGMS mission states, and you can easily find this on our website. It is that NIGMS is particularly interested in fundamental research. Basically, we are supporting-- basically, we are supporting basic research that aims to understand fundamental biological processes. Obviously, the long-term goal is to future advance disease diagnosis, treatment, and prevention, but our emphasis is not on curing a specific disease or tackling particular organ systems.
So if you're really focused on one specific question like that, that may fall out of the mission. So you should stick with our mission, which is fairly broad, and you can see it's hugely encompassing. You can find a lot of technologies that would fit this definition. Now, I did say that we don't want to go with a specific disease, which may tickle some of you who do clinical work or on a borderline that this may not be attractive. However, I would remind you that NIGMS does have interest in sepsis, anesthesiology, immunology, etcetera. So you can find, also, those clinically relevant areas defined on our website.
So you will still be able to come to us to seek maybe funding. Now, while you're developing your proposal and while you're thinking about your technology, whether it would be within the NIGMS mission, we strongly encourage you to reach out to us. We would be happy to discuss to see what you have, and you can chat with us to see whether that would be something that would fit. But in the next slide, please, I will show you-- please if you could move to next slide, Tony. Great. So these are potential technology areas for BTOD center.
And many of you are asking question, well, would my technology be a good fit? So one way to think about this is whether it fits the NIGMS mission. The other way is to follow, maybe, this list of potential technology areas. However, keep in mind as you read down these bullet points, this is not something that we would like to prescribe. We believe the scientists like yourself are the best to initiate the technology development, and you know which areas to go to the best.
And we would even encourage you to think outside of the box. You can look at this list and you see that there is a wide range of things that we are mentioning here. From bioanalytical chemistry and tools, we mentioned synthetic biology, high-throughput biochemistry, as you go down you see imaging tools, microfluidics, single cell technologies, a bunch of computational stuff. But we encourage you to use this as a guidelines, as a rail guide, not to stick with specific bullet item that we've listed in developing your own technology.
And also to come to us and talk to us. Like, what will you have? For instance, you may be thinking of developing a center that would come up with new methodologies in chemical synthesis, fine. NIGMS is a strong supporter of that. Talk to us.
If you're somebody working in synthetic biology like Dr. Ryan Mehl. We'll hear from him about his approach, but maybe you're thinking of engineering a new organism in which we can mass produce proteins, sure, let's talk about it. Or maybe you're thinking about biocatalysis and biomanufacturing and it's in embryo stages, we would be happy to hear about that. Or anything that you would be using maybe multiomics approaches at a single cell level. As you know, NIGMS funds research using a different magnifying glass. We look at the cell, we look at the organelles, we look at particular proteins, and we go down to atomic level resolution.
So we would be happy to hear from you. Next slide, please. And these are current BTOD centers that we are funding.
So these are six centers, and you can see that there are two that focus on NMR spectroscopy. They do different things. One in New York Structural Biology Center. The other one is down as a multi-institution National Resource for Advanced NMR Technology between Florida State University and the University of Florida. We also have a Center on Probes for Molecular Mechanotechnology at Emory in Atlanta.
We will hear today from Dr. Vicki Wysocki about her Native Mass Spectrometry Guided Structural Biology Center. This is something that is really hot, and the Ohio State University houses one of these centers. How to leverage and combine the native mass spectrometry with structural biology and maybe guide those complex structural biology questions.
Then we will hear from Dr. Ryan Mehl. I'm really happy that he's here with us presenting on his GCE4All center. This is really talking about how to utilize genetic code expansion and help a variety of biomedical and biochemical researchers across the state.
Dr. Ryan Mehl is at the Oregon State University heading his center. And then we have, also, Center on Cell Signaling Analysis. Again, multi-institutional program between UT Southwestern Medical Center and the University of North Carolina Chapel Hill.
Next slide, please. And without further ado, I would like to introduce our PI presenters today. And those are Dr. Ryan Mehl, as I mentioned,
who will present first on his GCE4All center. And then he will be followed by Dr. Vicki Wysocki from the Ohio State University, and she'll be talking about the Native Mass Spectrometry Center that guides structural biology efforts at the Ohio State University and across the nation.
Thank you very much. And, Ryan, why don't you take away? RYAN MEHL: Great. Thank you, Miljan, for the introduction. One thing that I can definitely say that was helpful for us in getting started and organizing our application was regular conversations with the program officers here.
That was something that really helped us understand the structure and the needs, so I would definitely advise anyone that is thinking about how to organize a center of this nature for this particular application to reach out and have multiple conversations. Because they are very big applications, and going down that road for a long length of time is annoying if you're off target. And I think that is something that should definitely be done by everyone that's going to be applying.
Go ahead to the next slide. Our center is called Genetic Code Expansion For All, or GCE4All. We are designed here to optimize and disseminate genetic code expansion technology, and I'll explain what that means. Our goal, really, is to take what this technology is and make it a basic tool that is accessible to all. And where we focus our energy is working with the DBPs to identify what are the roadblocks that they're having in advancing their NIH-funded research.
We pair them up with technology partners-- I mean, pair them up with our technology cores, and we have two cores in our center that I'll get into in a second. By working with them, we solve their problems, we optimize the technology to the level that suits their needs in order to publish their work. We then, obviously, do that with them, and at the same time with the optimized technology, we move that to the community engagement core of the center to get this out to broad dissemination. And so I'll get into what this is in a second. Let's go to the next slide. What is genetic code expansion? Most people have not necessarily heard of this technology.
It is essentially the ability to use translation to encode new amino acids into proteins inside living systems. This was first demonstrated back about 20 years ago. Since then, it has been demonstrated through many R21s and many R01s that you can put in hundreds and hundreds of different amino acids for many different applications. You can do this in many different model organisms. The general problem is that there are a lot of shortcomings when other people want to adapt the technology.
And so one of the concerns that we had when we were putting together this application is that we didn't see any centers out there that had-- that weren't focused around a major piece of equipment, right? There is no major piece of equipment for genetic code expansion, and we weren't sure how to handle that in our application. So this is a truly synthetic biology center. We are involved with figuring out how to optimize the synthesis of new amino acids, how to evolve new translation machinery to make it more improved, how to move into new organisms and optimize the technology in an organism that's needed by an NIH researcher. Oftentimes, the problem might just be scaling.
They just need a lot more protein to do the study in order to complete their project. Or how to alter the functionality of their particular amino acid of choice. So really for us, it is a multifaceted approach to synthetic-- I guess chemical biology and synthetic biology that most NIH researchers don't want to do on their own and so they come to us to get over that hurdle. So when we're done with all of that, obviously, our goal is to disseminate all of the optimized technologies to make this widely available. So I will now switch to a more visual side of that.
Let's go to the next slide. So in genetic code expansion, the way that we view this technology is here on the lower left. You have your amino acid. Oftentimes it's been demonstrated with one amino acid or another, maybe it's a post-translational modification, maybe it's a fluorescent amino acid.
The general idea is that sometimes it's not the right amino acid for moving forward with science, and that needs to be changed. Or someone has to figure out how to scale and produce that amino acid. Maybe that amino acid just isn't commercially available. So we try to identify what is the key shortcoming for any particular technology. The next step, of course, is evolving a tRNA synthetase for that amino acid.
Oftentimes in our technology there already is a particular tRNA synthetase, it just might not be good enough, or it doesn't work in the model organism at levels that are needed by the researcher. So we, obviously, will redo that as needed. And then, of course, at the end of this process, it's about making the protein.
How much protein are you making, is it pure, and how do you get past all of the impurities that might be problematic. And oftentimes it means going back and rebuilding the translational system to improve it so that the researcher isn't battling impurities that are complicating their science. One last step that is shown here with the red star is a lot of these amino acids are also reactive amino acids, and so people want to add labels to their protein. They don't have access to labels or the label that are being used are having challenges getting inside cells or washing out.
So there are label issues with regard to background or toxicity that we oftentimes overcome. So each different system is dealt with on its own merits, and we work with the DBPs to identify what system we need to target, and whether we can overcome those hurdles. So this is the overview of genetic code expansion technology. And so now I'm going to go to the next slide and talk about optimization. So one of the big things for us was-- there are hundreds and hundreds of amino acids, there's lots of translational systems, for us, it was really about first identifying, what are the core pieces that we think is going to be important to the NIH or NIH researchers? So of the hundreds of amino acid options out there, we decided to focus on two cores, or two top cores for technology optimization. One was bioorthogonal ligations because it would be broadly useful to a lot of our collaborators or driving biomedical projects.
That would enable work inside cells, it would allow you to label with lots of different fluorescent probes, spin labels, et cetera, for people to do work. Or you could also just purify those proteins and label them with in vitro for doing similar kinds of experiments. So that was one core. It's a very chemistry-oriented core. Most of the scientists that we work with in this area don't want to do any of that chemistry, they don't want to do any of that optimization, they just need it to work for their application.
And so that's how we picked the people that'll be associated with that science. Our other core had to do with what would be post-translational modifications or probes. So these are nonreactive amino acids, and they are used to interrogate biological function both in vivo and in vitro. And for that, we, obviously, picked-- we had to narrow it down to a key set of amino acids to stay focused. We focused on, initially, some amino acids that currently exist that just don't work well enough for NIH researchers to implement them broadly. And that was phosphorylation, acetylation.
When we get to probes, we minimize our set to fluorescent amino acids, fluorinated amino acids that could be used for pi-cation probes, as well as F19 amino acids that would be enable people to do F19, I guess, protein NMR. So our first goal was when we designed our center, we had to figure out what was the basis set that we thought would be most impactful for the NIH researchers. And then, of course, we had to have DBPs that were in need of those.
So it was a collaborative effort. We actually started with a lot more options, and then whittled this down to a much smaller set. Our current DBPs are listed here. They are all leading scientists that are funded by the NIH, doing their thing, trying to understand how biological systems work, and they just need our technology to work for their system.
So we pair up with them to identify exactly what is the linchpin, I guess, or the Achilles heel, if you will, for their application, and then work with them to overcome that goal. So that's how we organize the entire center. Why don't we go to the next slide? And as it was described by Miljan, really, all of these existed already as a proof of concept, but they just didn't work well enough for everybody to implement on their own. So for each system, there's a different shortcoming. And our goal was to identify, working with DBPs, what was that shortcoming, and then, we'll call it, kit-ify the technology and protocols so that they become really robust and they would work in other people's labs.
And so here is a really simple scenario-- and we'll go through some applications-- where the DBP gets the technology, and it doesn't work. And we troubleshoot why, we give them all the protocols, we give them all of the positive and negative controls to run, and we work with them to identify, why is this particular system not working in their hands, and then, of course, we rebuild it as need be. This, obviously, gets to the application.
They solve their problem. They publish their work. We now have optimized kits and protocols that we then push to our dissemination core. So that's the general process that was described by Miljan as well.
It works well for our system, our center. Let's go to the next slide. So here's some applications, and I cherrypicked just a few to focus.
One here is a fluorescent amino acid. James Peterson, one of our DBPs-- so one thing I do recommend when we talk about these DBPs and your technology is you really do want multiple DBPs using one technology so that you can really crossreference each other to the same technology, and you're getting multiple sets of feedback from DBPs on what's working and what's not working. We've had a lot of really great success where multiple DBPs are using a different-- or the same technology, and we can actually accelerate the proofreading or accelerate jumping through the hurdles that are presented to us by our users. Because we see different glitches from different people.
So James Peterson really wanted fluorescent amino acids with the bioorthogonal ligation, so dual encoding, so we had to, obviously, build that. Whereas Bill Zagotta and Sharona Gordon, they really just wanted the fluorescent amino acids to work in mammalian cells. So we rebuilt the chassis for a fluorescent amino acid that would be acceptable for both of them. I'm going to now describe one working scenario if we go to the next slide. So I would say, this is an example from Bill Zagotta and Sharona Gordon.
Before Genetic Code Expansion for All, before our center existed, they really wanted to be doing FRET on protein and inside cells. They were using a current amino acid that worked that they were using, which was called Anap. This amino acid really just didn't have the fluorescent lifetime requirements that they wanted. They didn't have the stability that they wanted.
And they knew that the Acd amino acid would, but they, obviously-- they had no access to it. So we built the genetic code expansion system for that that would work in mammalian cells. It improved their quantum yield by 2.5 times. It improved their photostability by 10-fold.
It allowed the first FLIM studies inside living cells. And, obviously, those improved fluorescent lifetimes were really critical for advancing their science. And that can be represented in that publication that we all worked to publish together. And through the center, you're going to have two different kinds of publications. You're going to have publications that are going to be led by your DBP, that are going to be publishing on the new front.
So here, they're the microscopists. They are the experts of studying proteins with their instrumentation. We are just giving them the tools to do so. That would be one paper that was led by the DBP. We clearly also have papers that are technology development papers where we lead those publications about how to improve the technology to get that out. So I think that's something to think about as you're designing your center, who's taking the lead on these different sides of the optimization cycle.
The next thing we did with them is, using the same technology, we were able to move to single exponential fluorescent lifetimes with Acd, and then using that system in order to do transition metal FRET. And that was a paper that was able to come out with that optimized system as well. So here is a system where simply just giving them access to the amino acid in a eukaryotic context that had the photostability and lifetime requirements to do their science opened up their ability to just push forward and get science done.
So that's one example. If we move to the next slide. With the same team, they also wanted to do a very different kind of technology besides FRET. They wanted to also be able to do DEER spectroscopy. And right now, one of the limitations about doing DEER spectroscopy in living cells was the EPR probes become quickly reduced inside a cellular environment.
What that meant for us is that if we were going to add a DEER or an EPR probe, not only did the EPR probe have to last longer, but also the labeling reaction had to be extremely fast so that it could be done-- that labeling reaction could be done before the DEER probe-- or the EPR probe was reduced inside the cellular environment. So working together with Bill, Sharona, and Stefan Stoll, who is the EPR expert, we optimized the encoding of a new amino acid, a tetrazine, which is shorter, which increased our labeling rate from what would be-- increased the labeling rate by about probably-- actually, I think that's on the next slide. So one second. If we just go to the next slide. The general problems here is that they came back to us and said, it's great that you might be able to label these amino acids, but if it's going to function as a DEER probe, it better be short because we really care about backbone conformational changes.
And so this is an area where they're telling you what's most important about the-- what to optimize. So the gold standard was a cysteine-labeled probe, really short, and labeled fast, but only worked in vitro. There were other in vitro labeling probes that worked slow like 400 micromolar per second and were quite long. So our job was to shorten that linker down and get the labeling rate up to something. In this case, we got it up to about 10 to the fifth. So we increased the labeling rate by a thousand-fold.
The next problem for us was to improve the stability of the label, and that's where we worked with Andrzej Rajca at University of Nebraska. So he's actually brought in as a technology partner. So as your center evolves, you can bring in people that would be not DBPs but they can be technology partners that have to solve a particular technology problem. Rajca here helped us with designing EPR probes that would last longer.
So if we can go to the next slide. Here is an example of the original probes in black and red, and you can see that their half-life inside cells was something in the order of 30 minutes to maybe an hour and a half. By going to a probe that he has designed that is in blue, we were able to push that timeline, we were able to push that out about 15 times longer. This enabled Stefan Stoll and Zagotta and Gordon to essentially complete the labeling in what would be about 15 minutes or less, and still gather all of the information they needed inside cells. And so this is an example of-- if we can go to the next slide.
This is an example where we published this paper, and we were the parent author because we had to build so much technology in order to make all of this happen. And now we're starting to apply that technology in their labs as a second round for optimization. So this is an example of where we are. We're constantly working with the DBP to refine bits and pieces of the GCE technology to make sure that it actually satisfies their requirements to move forward. If we can go to the next slide.
OK. So this is where-- just to summarize from those two examples, what we look for is, what do the NIH researchers need right now? Not in five or 10 or 15 years, but for the NIH researchers, it's like immediately what solves their R01 problem, if you will, right? The other way to look at this is, if you're building your center proposal, how do you have the maximum impact with the fewest research areas? This can balloon on you really easily to have lots of different research collaborators in lots of different areas. My recommendation would be to try to refine that to as few areas as possible with as many-- with multiple DBPs in those particular areas so that you're having a big impact. You can then roll those DBPs over and add a new area later, but the idea is that you really want to gain traction on solving a problem quickly for your DBPs. Again, I brought up, who drives the publication to completion? That's going to be split. Sometimes it's the center and your center staff.
Sometimes it's the DBP depending on the application. And that's just something to be mindful as you go into those collaborations. Another thing to think about as you're designing your center is how to cycle DBPs in and out of the center.
What's the lifetime? What are the checkpoints that say, OK, it's just not going to advance. This project isn't going to work out. We have to move on. Or, is there a natural six-month timeline? Is it a 12-month timeline? So those are things to think about as you design your center. When is your technology ready for broad distribution? This is always a tricky problem.
And so you can have optimization in cycles, but I would say the quicker you can get your technology out for broad distribution and feedback from the broad community, the better. So rolling out protocols and distribution of your technology sooner than later is probably the best thing to do. And if we can go to the next slide. And I'm just going to finish right here with our community engagement core. We have a number of different things that we're trying out, and it's never clear exactly what's going to be the best fit for your center. We do have a website and a knowledge base that provides a lot of the information and an interactive portal where people can contact us about future-- becoming a DBP, or about activities that are related to the center.
We have a training and support portal where we have a listserv set up to start developing question and answer sessions for people in the community as the community grows. We've started an international GCE webinar to get more people exposed to the technology so that they can understand how it can fit into the research. And that's the third Thursday of every month for about 3/4 of the year. We do have on-site workshops that we bring people in. This is great because what you get to do is you get to roll out your protocols that you've optimized with your DBPs.
We just had one where-- that was focused on bioorthogonal negations. And we had 20 people come in from around the world that needed a particular bioorthogonal ligation problem solved. We tested out the protocols with them in-house here for a week, and then we're able to send them home with kits that would work for them. So it gives you a lot of quick feedback on your dissemination protocols as well. In the context of dissemination, we are working-- since we are a synthetic biology-based center, we do work hand in hand with Addgene to set up web pages and disseminate a lot of our technology via plasmids or cell lines.
And so what is a protocol paired up with an Addgene web page to, in theory, give the researchers both sides of how to make that science work for them. And Addgene has been a great supporter of genetic code expansion technology, and we're working to make it really clear for what would be a global resource how GCE fits into solving problems, and how people can access the technology there. We also recognize that getting more people into GCE and getting industry people into GCE is important. So we do host-- we've been hosting conferences, and we'll be hosting another conference in 2024, where we're working to bring what would be developers and users together for the technology along with industrial partners that are trying to figure out how to gain traction in the technology. So it serves as a great brainstorming session for what should be future areas of research for the center. So with that, I think I'm going to stop.
I think that's the end of my slide deck. And I, obviously, will be around for questions. MILJAN SIMONOVIC: Thank you, Ryan. And we'll move to, Dr. Wysocki, please.
Thank you. VICKI WYSOCKI: Thank you. I'm happy to tell everyone about our Native Mass Spec RM1 at Ohio State and other locations. So this is a BTOD that is designed to develop native mass spectrometry tools to be used for structural biology, and to help perhaps guide other structural biology tools. As our main performance sites for the technology development, we have Ohio State, we have University of Michigan, West Virginia University, and Texas A&M. So we do have technology development going on in multiple sites, and then we try to share the technology among our different internal technology development sites as a first step of beta testing and making sure that the technologies are robust.
As has been mentioned-- next slide. So the heart of this program is having driving biomedical projects that present challenges and serve as test beds for our technology optimization projects. We do have three technology optimization projects which I'll tell you more about. Next slide. For our driving biomedical projects, we chose projects that would be both geographically and structurally diverse. So in developing native mass spectrometry, we didn't want to work on soluble easy proteins or small proteins only, we wanted to work on a range of different protein complex types so that people would see that these different technologies are applicable across a very broad swath of different kinds of structural biology.
And so we're working on everything from small soluble complexes, designed complexes, all the way up to large, very large complexes, RNA and DNA protein complexes, membrane protein complexes, heterogeneous glycoprotein complexes, and so on. Next slide. In addition to having the driving biomedical projects and the technology optimization projects, we, of course, have community engagement and sustainability as part of our projects, and I'll tell you a little bit about those as we go along. A lot of our dissemination involves vendors. So we have had as strong partners, Thermo Fisher, Waters, Bruker, Agilent, SCIEX, smaller companies. And on here, you'd also find-- if you were looking, you would find some additional universities that are not listed as part of our DBPs and not listed as a main technology development site.
And those are sites that are going to serve as Guinea pigs to move the technology into core facilities. So at our home locations, in particular, Ohio State, Michigan, and Texas A&M, we are using native mass spectrometry technology in core facility-type settings or trying to do that, and then we are also going to move it into some other sites to show them that they can either take existing instruments or get newer instruments and show their local investigators what can be done with native mass spectrometry. We find that people who haven't had exposure to this are really surprised at how much can be done with native mass spectrometry and immediately want access.
And we have a lot of people requesting to work with us, and we simply can't meet all the need for all of the people who would like to have a collaboration. So we're hoping to spread this much more widely. Next slide.
So our main goal is to optimize user-compatible native mass spectrometry technologies to characterize the assembly and disassembly of macromolecular protein complexes. That can include a lot of different kinds of tasks. So we could be measuring composition, stoichiometry, topology, stability, and flexibility, conformations, binding-induced changes, all kinds of different things. But there are some common challenges that need to be solved in order for this to happen for a wide variety of different protein complex types. Next slide.
So our technology optimization projects focus on the front end technology, where we are trying to separate components. Often, biological complexes of interest may be in a messy matrix, there may be other proteins present, other kinds of biomolecules present, we may need to control the temperature, we may need to raise the temperature if we want to try to intentionally unfold, for example. And so front end technologies are part of our TOP1, technology optimization 1. And technology optimization project 2, we are working on better selecting ions for fragmentation. How do we better separate them? How do we manipulate them, activate them? Do we use electrons? Do we collide them into a surface? Do we separate them using ion mobility technologies? And then in technology optimization project 3, we are working on technologies that look at ion stability.
And in both 2 and 3, we also have a computational component because to make this useful to a broad audience, we need for people to not have to know how to sit and manually interpret their data. The data types that can come out of these experiments can be complex, and we want software available that will take the native mass spectrometry data in conjunction with another structural biology tool, for example, maybe we have low-res cryo-EM, and we have an energy resolved surface-induced dissociation set of data, and we can use the two of those together to project and predict a quaternary structure of a complex. So we're working with-- Steffan Lindert from Ohio State is doing a lot of that computational work.
Next slide. And so I'm going to tell you just a couple of examples of some of the technology optimization projects, and explain the drivers for those, and why they're considered late stage enough to be part of this RM1 project. The first one I want to mention is something that we developed initially in our P41 period. But there is still a lot of optimization that needs to be done in the RM1, so it's appropriate for an RM1.
And that is automating the sample admission to the mass spec. So a lot of native mass spec is done by doing offline buffer exchange, by pulling capillaries, spraying from those pulled capillaries. We learned very quickly-- next slide. We learned very quickly that by working, for example, with David Baker at the Institute for Protein Design, they can make a lot of design protein complexes in a short amount of time, and we can't possibly look at all of those manually.
And if this moves into labs that are doing protein design, for example, they won't want to do this manually. So we developed a little column to separate salts from protein and just flow that into the mass spec, send the salt to waste. Seems like a really simple concept, but it hadn't been optimized. Next slide. When Thermo saw that we were working on this, they came along and said, we'd like to send people to visit, and we'd like to talk to you about developing a commercial version of this column.
And so this is considered late-stage technology at the moment because the first column is now commercially available. There's a Thermo Scientific NativePac OBE-1 Column. If you want to hear more about the development of this, there is a webinar that was sponsored by C&E News and Thermo that you can go and hear details of our development of this.
But that first column is now commercially available. It's been adopted by multiple labs for a lot of desalting-type chemistry that they want to do. Not necessarily even for protein complexes, but for some other applications. And we're now working on developing additional columns specifically for RNA separations. We can use this initial column for RNA, but we're looking to see if we can optimize that. We've also developed a 2D version, and we're looking at different column chemistries so that we can do an immunoaffinity purification as the first step and use that.
For example, we've shown that we can capture His-tagged protein complexes and let everything-- all the other proteins from an overexpression system go to waste, and then send the cleanup complex into the mass spec. So late stage because we've had some commercial success, but there's still a lot of optimization that needs to be done and driven really well by projects that require high-throughput. Next slide. And so another front end technology that we're working on-- and this is work from Dave Russell's lab but also present at the other-- at least, two of the other sites in our technology development center.
And that is technology to control the temperature of the spray so that we can look at melting curves for protein complexes, for example, and monitor those with mass spectrometry, measuring thermodynamic properties for protein complexes. There are a lot of biopharma applications where people are interested in measuring some of the thermodynamics of binding of a ligand, for example, to a complex. Next slide.
And so the types of projects that require this-- a lot of different projects would require this, but in particular, projects where we might want to be removing a ligand and looking at the protein unfolding, for example, or looking at the thermodynamics of binding of a particular ligand to a protein complex. Next slide. And so this is considered late stage because prototypes have been sent to multiple labs. There are several pharma collaborations going on right now, and it is a type of technology that can be considered multivendor.
It can be made universally available, it can be put on the front end of a mass spec, or a unique version of it can be specific to a particular vendor. But there's still a lot of optimization that needs to happen. Next slide.
Moving on to our technology optimization 2 projects. This is, sort of, once the ions are in the mass spec, what are some of the challenges that still need to be overcome? And one of those challenges is, how do we select at high mass to charge? Because these ions are being produced by electrospray ionization that produces a distribution of charged states, we like to be able to choose a particular charge state if we're going to fragment, and try to gain structural information, or define ligand binding, or define how different subunits are binding to each other, or what the overall topology is. We like to be able to select one charged state very cleanly. If there's a ligand involved, we don't want to select both the unligand bound, and ligand bound, or in this case, we have one ring of GroEL field and two rings of GroEL field.
We want to be able to select that very cleanly. And current quadrupole technology was sinusoidal waveforms doesn't do that as well as we would like. So we're developing, and this is led-- team lead on this is Dave Russell at Texas A&M. But they've developed a two quadrupole system where one of the quadrupoles can be used for cleaning up, knocking away detergent, for example. And the second quadrupole can have this digital waveform applied to very nicely select, a particular mass to charge of a particular protein complex so that it can then be sent for ion mobility, or fragmentation, mass measurement, so that we can get the maximum amount of information out of the complex. Next slide.
So this is considered late stage because we've illustrated it at both Texas A&M and Ohio State. So this is an example of developing the technology in one location, Texas A&M, and then shipping parts to Ohio State so that we can put this-- this is on the back end. I didn't really point that out, but it's on the back end of a commercial Orbitrap mass spectrometer so that we have a good analyzer to develop this technology with. We've illustrated, it's been illustrated by Dave in more than one vendor's mass spec, we don't have to change the quadrupole. We take the commercial quadrupole that's already in the instrument, but we apply a different power supply with a different waveform to do this kind of selection.
Next slide. Also, in our technology optimization 2 projects, we have this activation step. Once we've selected those ions, how do we activate them, and how do we maximize the information we get out of a particular complex? And so the example I'm showing you here is spike protein, spike proteins prepared.
This is a particular stabilized spike protein provided by Erica Ollmann Saphire's lab from the La Jolla Institute for Immunology. So think of this as part of a vaccine development project. This is being produced in such a way in cells where we will have a heterogeneous distribution of glycosylation. So these gold pieces are glycans attached to this spike protein, which is stabilized by disulfide bonds. So the three different monomers of this spike protein, SARS-CoV-2 are covalently bound to each other to stabilize the spike trimer.
And it's a HexaPro construct so that this is a very stable spike protein. But when you try to spray that in a normal native mass spectrometry experiment, you get a big blobby peak, you don't get resolved charged states because of the heterogeneity from the glycosylation. And so what we can do is we can use quadrupole mass selection to mass select a thin slice of that peak.
We can then allow those ions to pass through a filament region where we're generating electrons. Those electrons’ charge reduce the complex, and we get a resolved distribution. So we can make a much better mass resolved spectrum than we would get from just this big broad distribution that you see here.
That technology-- next slide-- is being used not just for this type of project, but also in some of our collaborating service projects with people who make adeno-associated virus capsids. The surface-induced dissociation that we can do-- and I didn't point out, but this little device that we've installed into the Orbitrap mass spectrometer to do the electron capture also has a device that we developed to do surface-induced dissociation, where we select ions and collide them into a surface. And when we collide AAVs into this surface, we get extensive fragmentation into substructural pieces.
This is hard to do. If you just collide into a target gas, that would be used in a normal commercially-available tandem mass spectrometers that we get. By using the electron capture coupled with the surface-induced dissociation, we can pull apart the charged state distributions that are part of a particular fragment like this 15-mer that's part of this capsid.
That 15-mer, we can look at its fragments. This is a charge detection mass spectrum where we have mass to charge and we have mass on the y-axis. And we can see that the viral proteins making up this 15-mer peak, for example, are a range of different compositions of VP, viral protein 3, 2, and 1, the three main viral proteins that make up this capsid. It's really hard to get this information by any other means.
And so working on technology to try to make this possible for people to better characterize things like heterogeneous glycoproteins or capsid types protein complexes. Next slide. In our technology optimization project 3, we are using that surface collisions that I talked about, and also gas collisions that are typical in commercial mass spectrometers to look at protein unfolding and protein stability. What are the different intermediates formed when a particular protein unfolds, for example? A lot of the development of this has been done by Brandon Ruotolo at Michigan.
And what we've started to show is that by having our surface collisions, we can also cause some access to fragmentation patterns that are not accessible by these low-mass target gases, and so better characterize some of these larger and more complex systems or some of the systems that are more stable for different reasons. Next slide. And so we consider this late stage because the collision-induced unfolding option is actually now adopted in dozens of labs worldwide. There are many pharma collaborators. There's multivendor adoption starting to occur.
But there's still pieces of this technology that need to be worked out. The surface collision part needs to be worked out. Next. Last one that I will show you. Technology optimization 2 and 3, as I mentioned, have computational components, and those include integrative modeling for quaternary structure, where we combine the mass spec and ion mobility data, for example, with complementary low-resolution structural data from other structural biology tools to come up with better structures.
Next slide. This is late stage because as these tools are developed and are being beta tested, we start providing them immediately in Rosetta, for example, or in OpenFold format so that people can use them. And final slide. Next slide. Oops, sorry. Next to last slide.
We do have two technology development-- go forward, please. We do have two technology development projects. I won't dwell on those, but just finish with our community engagement. We have our website where you can find an overview of projects, protocols, a contact link if you want to do work with us. We are converting this over currently from our P41 website to our RM1 website, but a lot of the protocols are valid and useful. Our training and support involves short courses, workshops, webinars.
On-site and partner training, we think it's useful if our staff can go to our partner sites if they are trying to adopt a technology and do some of that training. We're doing a lot of collaborator training because in the end, we want the actual biological users, the structural biologists to be able to use the techniques and choose the proper flavors in their labs. Our dissemination is involving vendor products, prototypes to labs, software development that we make available, publications and presentations. We think our graduate placements are important. We've graduated a lot of students who have worked on these projects, and they then carry a lot of the technology forward with them to their companies or to their academic positions. We have instituted, we're just instituting for our RM1 an industrial advisory board so that we get more pharma and instrument company input.
And as I mentioned, we're moving the technology into core facilities. So I'll stop there, and I'll be available for questions. DOROTHY BECKETT: So I'd like to thank both of our presenters. And please feel free to enter any questions that you have into the chat box. I'm going to start with a few questions for the two PIs, and this is related, also, to some of the questions that we've seen in the chat box.
So the first is, how was your technology development supported prior to establishing the center? VICKI WYSOCKI: I'm happy to start answering because I just noticed I still have my microphone open. We had a variety of ways. In the early days, some of the development of devices was being done with NSF funding, R01 funding.
Some of it for our RM1 was done in our P41. So a whole variety of different funding mechanisms were used to bring technology to the stage where it was ready to go into the RM1. DOROTHY BECKETT: And, Ryan? RYAN MEHL: For us, I guess we'll call it our preliminary results for our center, I think, happened through a number of collaborations that happen organically when you're optimizing technology like this. We were able to secure a technology focused R01 that did enable us to showcase that we had a workflow that I think was amenable to our center-- the center mission, right? But that was-- besides that, those are the primary, I guess, mechanisms. One, just collaborating with people to solve problems to demonstrate that it was possible, and two, a technology-based R01 that helped us get over the hurdle. DOROTHY BECKETT: I think I'm going to take a question for both PIs from the audience or the attendees, Mei Hong has this question, to both centers, how many staff do you have to support the DBP and CE activities, and are these staff mostly funded by the RM1? RYAN MEHL: I'll say quickly.
For us, I think all of that's going to be related to how many-- so the budget that you're going to get from NIH, I don't know how flexible that is, but I think it's a fairly defined budget range. And so, again, you're going to have a certain number of technology cores and a certain number of DBPs. And so the more times you have a technology core that can, say, serve five DBPs, the more you're going to be able to do with less on-ground effort, if you will. You might have a lot more meetings with all of those people, but you don't necessarily have to reinvent-- you're not doing separate technology developments for each of your DBPs. So for us, I would say that we definitely have, like, how many staff, right? It's hard for me to say-- I would say that we have senior staff as well as people that are in the trenches in the lab. Right now we probably have two to three people per core at a minimum that are central, that are leading each of the cores.
VICKI WYSOCKI: Yeah. We tend to use a lot of grad students and postdocs and don't have as many senior staff on this. Everything is determined by the max budget or the budget that you've received, and it's a very fixed budget. We've tried to expand what we can do by having some synergistic grants. We were fortunate to have a-- participate in a couple of SBIRs, for example.
And that can help take part of the load off, and then we can focus on some of the technology optimization by taking some of the work off into other funding. DOROTHY BECKETT: I'm going to ask one more question that we had actually prepared that we thought the group would be interested in is, so please comment on the balance. How do you balance directing your own research activities with directing the center? VICKI WYSOCKI: Being organized and holding lots of meetings. I'm not sure if that's what you're looking for. The center is part of my research activity is part of the answer, right? And then with other projects that we have, just dividing up time.
And I believe in lots of collaborator meetings, so we have lots of meetings with collaborators and colleagues for each project, and we keep them all moving in that way. RYAN MEHL: For me-- I think Vicki is probably a bit more experienced at running centers. This is my first step into that space, and so I'm probably maybe a little bit more overwhelmed by the whole thing. I do have R01 funding, and I do have NSF funding, and so I have other projects that are running. We also have other collaborative projects that we're doing at the same time. It's really-- it's hard.
The center is your research, right? So you have to really decide that optimizing and disseminating this kind of science is your goal, and that is something that really matters to you, and that is something you genuinely really want to do. And that becomes your research, and that's an important thing to make sure is in the right space for you emotionally before you go down that road. Because if making this work for other people and disseminating it is not part of who you are, this will just be awful. VICKI WYSOCKI: And I would echo that because a program officer actually said to me at the time that I said, hey, I think I want to turn in one of these center grants, they said, you realize this is going to be very different from all the R01s that you've always led.
This is a very different kind of science, and you have to be aware of that. And I want to add one other thing. One reason that we are able to get a lot done in our center is because we have an amazing admin person, Laura, who's probably on the call today or on this webinar. And she is just amazing. And so one of the things that I think has also been important to us is we have asked the partner universities, the universities who have technology development going on to also support us with some matching funds, which has allowed us to do things like pay for some administrative support, and to extend the number of grad students that can work on the projects because we get fee waivers for tuition and those kinds of things. So I think, for us, one way that we have magnified the money is to have some local funds that are also contributing from each of our partner universities.
DOROTHY BECKETT: So thanks, Vicki and Ryan. Your answer to that last question actually answered a lot of other questions-- a few other questions that we have, what motivated you, and it's clear that the motivation is that you want to get this technology out to the community. OK.
I think we're going to go to some questions that the NIGMS staff can answer. So one of the questions I'll ask you, Alvin, is it OK if the proof of concept or prototype are funded by other agencies and not the NIH? ALVIN YEH: Yes. I guess the answer would be, yes.
That's no, there's no requirement that the technology be developed with support from NIH. I think this question of the stage of the technology, ultimately, I think it's probably up to you to convince the reviewers that it is at the appropriate stage, in my mind, my personal mind, that this would be a technology that was useful for you in the lab. It needs to be, it can only be, or it's very difficult for nonexperts to use, it's really at that stage where experts can use it only, and
2023-10-16 22:34


