MIT.nano Seminar Series: Genevieve Van de Bittner, PhD
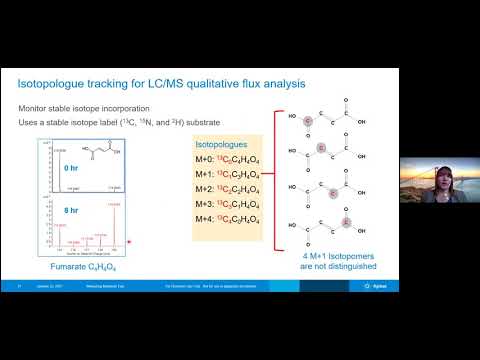
good afternoon everyone uh and happy new year welcome to our first seminar of 2021. uh it is my pleasure to introduce today's speaker dr genevieve van de bedner who is joining us from agilent research laboratories genevieve joined agility in 2016 with the goal of improving measurement technologies prior to that she completed her phd at uc berkeley and completed her postdoctoral research at harvard medical school at agile at agilent dr van de binner's work focuses on developing automated sample preparation methods for lcms metabolomic and lipodomic analyses and she also explores the combination of seahorse and mass spectrometry metabolomic data so it is our pleasure to have genevieve here with us today to learn more about measuring metabolic flux with cellular and molecular resolution before i turn it over to genevieve i would just like to remind everyone that you can ask your questions at the end of the seminar either by typing it using the q a feature of this webinar or you can also raise your hand and directly ask your questions so with that genevieve thank you very much for being here today we look forward to hearing your talk and i turn it over to you thank you farnaz all right so today as farnaz indicated i'll be talking a little bit about measuring metabolic flux or measuring the rates of metabolic reactions and doing this at both cellular and molecular resolution and in order to do these types of measurements i'll be sharing how we use a sea horse xf and lcms qualitative flux analysis technologies so to first give a little bit of background on why metabolic flux measurements can be so important i wanted to direct you to what common cell analysis technologies typically do for cell analysis they often take a steady state or end point measurement and this really gives us a view of of the cells that is like a photograph so we get a steady state view in this example i'm showing on this slide you might have a highway with cars on it and the information you're getting from this type of analysis is really just the number of cars in a given section of the highway at a given time on the other hand in a metabolic flux study you are getting information that's more akin to a video so we're really um able to zero in on things like how fast are all of these five cars moving on the highway are they moving slower or are they moving faster and by getting this sorts uh this sort of information we're able to know the rate of the traffic and this can provide insights into where there are traffic jams in cells this might be a regions where there aren't enzymes that are inhibited or mutated or in low abundance we can also get an idea of where traffic is moving freely at normal speed and this might be uh give an indication of where enzymes are functioning normally and there are normal concentrations of enzymes as well as the metabolites that they're interconverting um and then on the other hand we might find regions where there's really slow traffic and the rate's really slow and uh or sorry where the the rate is really fast and this might indicate uh areas where we have upregulated enzymes um or gain of function enzymes uh for instance so um the seahorse and the lcms qualitative flux analyses allow us to get these sort of rate measurements and better understand um how fast metabolic inter interconversions are happening within cells but they do this at two different resolutions so the sea force xf analyses really provide us an idea of what our metabolic flux or metabolic rates are at a more city scale view so what is the combined rate of cars into and out of a city on the other hand the lcms qualitative flux analysis really provides us measurements that are more at the street scale so we're getting more information about what are the rate of cars on each of the individual streets within the city in other words a molecular resolution view um so we'll take a deeper dive into both of these technologies but um essentially what the seahorse xf analysis is giving us is a view into uh the cellular resolution of how metabolic flux is happening um within cells sort of at a global scale and the sea horse accept all technologies focus on looking at glycolysis on one hand and then respiration on the other and i'll go into some details on that um and then the lcms qualitative flux analyses are really providing us molecular resolution information of how metabolites are interconverting within the cells and at what speeds they're doing this so this is really at the molecular scale so this is just a view of the two technologies and what they look like um so i'll focus on the seahorse xf technologies first and that's shown on the left so again the seahorse xf uh xf measurements are done at sort of a cellular scale and telling us globally what cells are doing and the xf assays measure the rates of change of two key energy metabolism pathways the first pathway is again glycolysis and in particular for the glycolysis measurements what we're monitoring is we're actually monitoring the extracellular acidification rate um that is being caused by the cell's metabolism so in particular cells will pump lactate into the extracellular space and we can monitor this lactate production by looking at changes in ph on the other hand uh the seahorse technologies allow us to also look at respiration and in the case of respiration what we're really looking at is we're looking at oxygen consumption by the cells so this is going to be looking at the oxygen consumption rate of the cells and really here you're looking at how much are the mitochondria within the cells using oxygen and funneling that through the electron transport chain to make atp so an important note about the seahorse assays is that during our ecar and oh car analyses there are four injection ports that we can use and these can be used to add different compounds to the cells and then we can look at what the metabolic impact of these compounds are and we use that to develop a lot of different um test kits that allow us to look at different components of glycolysis and mitochondrial respiration so in the first test kit i'll i'll just go over a couple of these so you're familiar with them before i go into the real data of telling what cells are doing under different conditions but in one of our our test kits we have the cell mitostrus test kit and what this does is it really focuses on the oxygen consumption of the cells or that o car value and we're able to inject different compounds including oligomycin fccp and anetomycin a and wrote known to the cells during this assay the cells are live and we can monitor what happens once these compounds are injected and these compounds do different things to the cells and allow us to monitor um for instance the atp production by the mitochondrial by mitochondrial respiration we can also get an idea for what is the maximal capacity of the cells uh to consume oxygen um and then we can also look at the basal respiration levels and non-mitochondrial respiration in one of our other kits we have the glycolysis stress test kit and in this kit we can look and zoom in more on the glycolysis or the extracellular acidification ecar rates and in this case we again inject compounds to the cells um as they're live and you know going on their regular business and we can use these compounds to detect what is uh the normal basal rate of glycolysis in these cells um what is the maximal glycolytic capacity in the cells so if they really were pushed to the extreme how much um how much flux is there through the glycolytic pathway um and then we also get a measurement for non-glycolytic acidification of the extracellular media another one of the test kits that will come up in the first example i'll show later is the cell energy phenotype test and in this test what we're doing is we're focusing on both oxygen consumption rate as well as extracellular acidification rate and as we look at both of these measurements we put the cells under stress so we can see what their baseline okar and ecar are as well as what their stress okar and ecar are in the next assay we have a real-time atp rate assay and this assay really zeroes in on what is the amount of atp rate atp produced by both mitochondrial respiration and glycolysis so this is an atp production rate assay and again zeroing in on both the respiration and glycolysis of the cells and then finally another uh kit that will come up in one of my examples is the substrate oxidation stress test and this is very similar to the cell midas stress test um but in this particular case we're going to be able to look at how much the cells are using different fuel substrates to fuel their oxygen consumption so we can in this test look at whether cells are using long chain fatty acids glucose and pyruvate or glutamine to really fuel their oxygen consumption and their electron transport chain okay so that's a little bit of background on the sea horse assays and next i wanted to give some background into the lcms qualitative flux analysis in this type of analysis what we're doing is we're using isotope labeled metabolites to really understand what are the rates of interconversion of metabolites so we can add these isotopically labeled metabolites to cell culture we can let them sit in that cell culture let them be metabolized by the cells and then here we use the lcms technology to both separate those metabolites out in liquid chromatography and then we can measure the mass of those metabolites using mass spectrometry and this really helps us isolate the different isotopes that were in the cells what we added to the cells and then also where these isotope went isotopes went once they were introduced to the cells through some software that we have the agilent vistaflux software we're able to analyze these isotopes and and pull them out of the samples that we analyzed and then we can see where those isotopic labels ended up and then we also have a visualization software where we can take the metabolites found in vistaflux and map those onto metabolic pathways so you get a real view of where the isotope labels are going and what pathways they're involved in so just to give a slightly deeper dive on the qualitative flux analysis i wanted to go over isotope log tracking so what we're doing in our our analyses here is we're tracking isotopologs and we're gonna have an example of fumarate that's shown on the left here so in fumarate we have four different carbons if we incubate cells with a carbon 13 labeled glucose for example this will eventually get into labeled interfumarate and this will so the glucose goes through glycolysis and then goes into the mitochondria and it goes into the tca cycle and one of the metabolites in the tca cycle is fumarate so we can get that label from glucose into fumarate and we can see in our mass spectrometry data fumarate in in the case where it's unlabeled on the left here and then as each carbon 12 is converted into a carbon 13 we can start to see those mass peaks for each additional carbon added and when we uh talk about isotopologs we're really talking about combining all the isotopomers of of a given mass so in fumarate if we have an m plus one mass so there's one carbon 13 label that carbon 13 label can be at any position within this fumarate molecule and we just combine all of those together in our particular analysis and so that means we're looking at isotopologs instead of the specific isotopomers to look at the isotopomers you would need to have fragmentation and it's something that you can do but is not typically done in this qualitative flux analysis um so that's a little more information about the labeling and then to talk a little bit about the software and how it works before the experiment is done we actually decide which metabolites we want to look for and so this would be helped by taking maybe your metabolic pathways of interest uh making a list of all of the metabolites in those pathways and then knowing the retention times for your particular um liquid chromatography method and so we put all of those into the software and then the software is able to find uh the compounds for the particular masses of the compounds we can look at the mass spectra and then we also get a picture of our isotopology so in this group up here we have no label no carbon 13 labeling in this group um and down at the bottom we see there's some of this m plus zero or non-carbon-13 labeled molecule and then on the right we also have a fully labeled um carbon-13 molecule again this is a fumarate example once we have this data and we know um how much of a compound is labeled in any given sample we can lay that onto this metabolic pathway using a pathway mapping software so in this case we can again look at fumarate down here and look at how it's labeled across those different samples shown on the previous slide okay so now that i've given a little bit of background on both of these technologies i want to cover a couple different examples um and and actually share with you how can what can these technologies help us understand um there are a lot of different things that cells can do and they can rewire their metabolism to change their function and this is often in response to some sort of external stimuli and when cells change their function some of the functional changes that can happen include activation of cells uh proliferation of cells differentiation apoptosis and dysfunction and so we'll go over some examples that show how cells are changing function and how we can monitor how metabolic rates are changing during this change in in cell function and the examples that i'll be covering in this particular talk include looking at um the activation of macrophages um the cool temperature adaptation of adipocyte or fat cells and then we'll also look at fuel sources uh for cancer cancer cells okay so the first example i'll focus on metabolism in macrophages in particular when they're undergoing activation and again we're going to look at this with both cellular and molecular resolution so both the sea horse and lcms resolution so to give you a general view of what is going on in these cells just as a primer for this example we have macrophage cells again as i mentioned on the last slide and we're really in this example going to focus in on glycolysis and the rates of metabolites through the glycolytic pathway and then we'll also take a look at the tca cycle and the rates of metabolites through the tca cycle which is again correlated with the oxygen consumption of the cells and again the glycolysis is correlated with the extracellular acidification of the cells the data that i'll be showing for this example was provided by professor gerald uh leroy mahmoud from the imperial college of london and so we thank him for his help with that okay so just to go over the design of this of this assay again we're going to be looking at macrophages um a type of immune cell and these ac these macrophages are activated with a compound called lipopolysaccharides or lps these lipid polysaccharides are produced by bacteria and so when cell immune cells see these lipopolysaccharide they kind of say hey there's um there's a danger here we need to activate and we need to produce an immune response so in order to confirm the activation of these macrophage cells by lps you can look for a tnf alpha production and tnf alpha is a cytokine that these macrophages that these macrophages produce when when they are uh interact with little polysaccharides or when they're activated so we can see in the lps treated case um the macrophages do indeed produce more tnf alpha and then if we do our cellular resolution analysis of metabolic rates in these cells um again we're doing that we're looking at the cell phenotype assay and we're looking at both oxygen consumption rate and extracellular acidification rate of these cells and in we also look at these cells under a basal condition as well as a stress condition and what we can see is that the lps treated cells have increased extracellular acidification or increased glycolytic rates under both the basal and the stress conditions and then the lps treated cells also have less ability or less maximal oxygen consumption rate than the untreated cells so once we've looked at the cellular resolution and seen the changes in glycolysis and ocar at a cellular resolution we can then take this down to the molecular resolution and do our lcms uh qualitative flux analysis so in this particular case again we're focusing on glycolysis and the tca cycle and we can look at all of them we can pull out all of the metabolites from both of these pathways and look for them during our lcms analysis in this particular analysis um it was itaconic acid was also looked at so we'll see that so in this uh lcms qualitative flux analysis uh ubiquitously labeled carbon 13 uh glucose was added to the cells and it was added to both the control and lps activated macrophages and this carbon 13 glucose was able to go through the different metabolic pathways within the cells and we were able to look at the incorporation into the different metabolites and so what we can see from our pathway map here is that for some of the key glycolysis glycolysis intermediates we do see an increase in carbon 13 label incorporation into those metabolites so for instance this glyceraldehyde 3-phosphate or also looking at lactate over here if we look at the tca cycle what we notice is that there's actually a decrease in metabolites found in the reductive branch of the tca cycle so there's a decrease over here but there's an increase in carbon 13 labeling of the metabolites on the oxidative branch of the tca cycle and here's where etaconic acid becomes important to look at because it was found that although glycolysis is increased in these cells and that glycolysis can lead into the tca cycle what was happening in the tca cycle is these metabolites were going through part of the cycle but then being funneled out of it and and into idaconic acid so this additional molecular resolution showed us exactly what was happening to those different metabolites in the second example i'll be covering uh the fuel sources for primary adipocytes and what happens to metabolic rates during cool temperature adaptation of these cells so this work was uh provided and the data was provided by professor orman macdougald at the university of michigan in this case we have a similar view of the cell but keep in mind we're now looking at adipocyte cells or fat cells so we have some additional components in here we'll look at glycolysis again as well as the tca cycle but in this case we'll also take a look at the pentose phosphate shunt um as well as lipolysis again these are fat cells and that can easily undergo lipolysis as well as beta oxidation and again we're looking at what happens to these fat cells when they are subjected to a cool temperature over long periods of time so in this particular analysis there was some rna-seq done and in the rna-seq data it was expected that during cool temperature adaptation of these cells that oxidative phosphorylation would actually be increasing so this is the reverse of the previous example and it was expected that glycolysis would be decreasing in these cells again a reverse of the previous example if we take a look at the oxygen consumption rate of these cells and we look at the 31 degrees celsius culture we can see that the basal background oxygen consumption rate of these cells has increased versus cells kept at 37 degrees celsius and then again in this section here we also see that there's an increased oxidative capacity of these cells so their maximal oxygen consumption rate is also much higher than the 37 degrees c cultured cells we can also use the rna seq analysis to look more specifically at different genes and here we see that in addition to having a reduced glycolysis in these cells um shown here it's also expected that cool adaptation would reduce the flux through the pentose phosphate shunt as well as through amino acid biosynthesis and glutathione metabolism so in order to monitor these other different pathways we're now again able to do this lcms qualitative flux analysis and actually look at how carbon 13 labels are incorporated into specific metabolites so in this particular case there were three different isotope labels that were used um the carbon 13 labeled glucose carbon 13 labeled pyruvate and carbon 13 labeled glutamine and so this is just showing the pathway again on the left at the top we'll focus on the carbon 13 labeled glucose data and what we see here is that we again confirm this reduced flux of glucose through the glycolysis pathway by looking at carbon-13 labeled glucose we see at 37 degrees there's quite a bit of labeling that gets incorporated but at 31 degrees this labeling is much reduced and you can see that um not only for fructose 6-phosphate and glucose 6-phosphate but also for some other metabolites in that glycolytic pathway if we take a look at the pentose phosphate shunt we also see that there's a decrease in label incorporation of glucose into ribulose and cellulose 5-phosphate so again we're seeing a reduction in the rate through of glucose through that pathway and then if we take a look at the tca cycle if we look at citrate or isocitrate as well as malate we again see this reduction in labeling of the pathway when the cells are at 31 degrees celsius so if we take a look uh now at pyruvate and glutamine we find a sort of reverse picture here and we find that at 31 degrees celsius the labels from pyruvate and glutamine actually are incorporated more in those cells at the cool temperatures than at the at the higher temperature but does this really explain all of the increase in oxygen consumption rate that we originally saw with the sea horse assay so again we went back or they went back to the sea horse assay and decided to look at the oxygen consumption rate in more detail and at this time we're using uh an assay where we're able to look at the fuel sources of the oxygen consumption rate and so in this particular case okar experiments were completed with inhibitors of mitochondrial fatty acid uptake as well as inhibitors of lipolysis so when we add this inhibitor of the fatty acid uptake into the mitochondria so we're adding atomoxer in this case we see that this increased oxygen consumption rate at cool temperatures is actually reduced back to this sort of normal 37 degree case um and then if we also use this lipolysis inhibitor we again see a reduction in this increase oxygen consumption rate and so both of these together indicate that the oxygen consumption rate increase in cool temperature adaptation is really coming by coming from uh lipolysis and beta oxidation of fatty acids in the mitochondria so in this particular story we're seeing how transcriptomic seahorse xf and lcms flux analysis all really combine to reveal what is happening in the metabolic rates of these fat cells that are adapted to 31 degrees celsius all right so i'll get on to my next example here and i'm trying to see if i can get rid of this bar at the top but it doesn't look like it's going away um okay so in the third example i'll be covering some fuel sources for different cancer cells and again we're going to look at this at both cellular and molecular resolution okay so in this particular example we'll be looking at the metabolic phenotypes of non-small cell lung cancer cells and it is well shown in the literature that there are some predominant mutations that are found in some of the non-small cell lung cancer cells in particular there's this k-ras mutant that happens in about a quarter of these types of cancers and then we have the egfr mutant that happens in about 20 20 of these cells so in the non-small cell lung cancer um there there is some uh expectation that some of these mutations will have an impact on cellular metabolic phenotype and in particular there's some suggestion that the some of these cells may be using lactate as a major fuel source and so we wanted to study this a little bit deeper in this particular example again we'll have a large focus on glycolysis in the tca cycle and the electron transport chain but we're also going to focus in on uh lactate transport in these cells so in in particular we're focusing on two different mutants as i mentioned before we have a pc9 mutant for the egfr mutation and an a549 mutant for the k-ras mutation and these cells are going to be fueled with both glucose and lactate and then we'll take a look at inhibition of some enzymes that are involved or thought to be involved in this lactate transport so in particular we'll be looking at an inhibitor for the mitochondrial pyruvate carrier so when lactate enters the cell it's converted it can be converted into pyruvate and then that can be carried into the mitochondria to fuel the tca cycle and we'll also look at inhibition of the monocarboxylate transporter one which is uh well known in many cell examples to transport lactate into and out of the cell so in this particular analysis for the seahorse portion of the analysis we focused on the real-time atp rate assay and again this assay really tells us what component of atp production is being used to fuel um uh is being fueled by mitochondria versus being fueled by glycolysis and there was actually a study done uh internally to agilent where we looked at a couple of different cancer cell lines and tried to look at uh how mitochondrial atp production compared to glycolytic atp atp production in these cells and we found some maybe surprising results um so we found that there are cancer cells that heavily fuel their atp production by mitochondrial respiration and others that really heavily fuel their their atp production through glycolysis but there wasn't necessarily an inverse relationship here so there are some cells that have very little atp production overall and that was interesting to see back to our non-small cell lung cancer example um here we're showing the pc9 mutant egfr mutant cells on the left and then the k-ras mutant cells on the right and here again we're going to be looking at cells that are fueled with both lactate and glucose and what we found from these particular studies is that the egfr egfr mutated cells actually use lactate to increase their mitochondrial atp production so they're shuttling lactate into the cell and then using that lactate as a fuel source on the other hand the k-ras mutant didn't seem to be using lactate in this way so although these are both two types of non-small cell non-small cell lung cancer cancers they're actually functioning in different ways metabolically so if we take a deeper dive into the pc9 cells and look at what is actually helping the cells use lactate for energy we did our inhibition studies here so we're inhibiting mct 1 in the red boxes and then inhibiting mpc in the in the purple boxes and what we found here is that the inhibition of mct-1 does not actually reduce the amount of mitochondrial atp production but inhibition of mpc did reduce that and so this was interesting because again mct-1 is the known lactate transporter but in this particular case it doesn't seem to be helping with the import of lactate into the cells for the fuel utilization we also did a qualitative flux analysis to get a more detailed view of what was happening to specific metabolites in the tca cycle um and and seeing uh how lactate was being incorporated into these metabolites so again we're looking at the pce 9 cells and we're looking at uh addition of carbon 13 labeled lactate to these cells so we have lactate box up here and then succinate box down here and we've also taken the data from the previous slide and sort of rotated it so that it's easy to compare with this metabolic flux data but what we see here is there's actually quite good comparison between this data so if we take these four conditions here um they actually map pretty well with uh these four bars from the qualitative flux analysis um so what we see is that in in the case of the vehicle or the mct-1 inhibition we do see high incorporation of this labeled lactate into all of these tca cycle intermediates but once we add that mpc inhib inhibit inhibitor again so the uk in these two cases of that addition we actually see a much lower uh flow of lactate into these tca cycle intermediates okay so the kind of conclusion from this study is that the cellular mitochondrial atp production is really resulting from molecular uh flux of lactate through the tca cycle and again we can take a deeper dive into what's happening to lactate in succinate in particular um so in the lactate on the left here since we're adding carbon uh ubiquitously labeled carbon carbon-13 lactate we expect a high amount of this m plus three labeled lactate to be present and then for the succinate we can take a view of this over here and what we expect in in our case a vehicle only is that we would see incorporation of this lactate label into succinate in particular there's a slightly higher amount of this m plus two succinate um this makes a lot of sense from a tca cycle standpoint because in that first turn of the tca cycle we're going to incr incorporate two carbons from lactate into succinate um and so this is the highest peak and then on subsequent turns of the tca cycle will actually incorporate more carbon 13 labeling all right so yeah i'm speeding through these examples but i did want to get to this last example of inhibiting fuel usage by cancer cells and looking at this again with the cellular and molecular resolution so in this particular example we're going to again be taking a look at cancer cells and in this case we'll be looking at the inhibition of oxidative phosphorylation and what this does to all sorts of metabolic pathways within the cells and the rates of metabolism through them so this data comes from uh pietro morlocky and jennifer molina who completed the studies at the md can andy md anderson cancer center um in this particular case we'll again focus somewhat on glycolysis and the tca cycle but we will also take a look at again the pentose phosphate shunt and the production of nucleotides as well as taking a look at aspartate and the production of pyrimidines from aspartate um so these are just a couple of the reasons why inhibition of oxidative phosphorylation is of interest for cancer there are some cancer cells that are heavily dependent on oxidative phosphorylation for production of their energy and one of these cancer types is acute myeloid leukemia there's also some data in the literature suggesting that tumors can become more sensitive to inhibition of oxidative phosphorylation after the standard of care treatment so there might be some room for having combination therapeutics with an oxidative phosphorylation inhibitor so uh the md anderson cancer center cancer center went through a med chem screening to look for these oxford loss inhibitors and they were able to find this compound iacs 10759 that actually inhibits complex one of the electron transport chain and this particular compound had a lot of favorable qualities like it had a low nanomolar activity in vitro it was also low enough in toxicity and had a good pharmacokinetic profile so all of these combined to make them choose and study this compound in a little more detail so in this particular case as expected when uh acute myeloid leukemia cells are incubated with this iacs inhibitor we actually find that there's a decrease in baseline oxygen consumption rates this is completely expected we're inhibiting one of the enzymes of the electron transport chain so definitely our consumption of oxygen is going to go down in these cells and it does so in a dose-dependent manner what we see um that happens or is the result of this is that there's an increase in cell death as well as reduced viability and reduced cell proliferation after in inhibition of this complex one of the electron transport chain picture cellular resolution view of what's going on the real question was what is actually happening in these cells and which metabolites are impacted the most um and so in this particular example there was a targeted metabolomics analysis of 80 different metabolites and this was just a more steady state analysis of looking at after we treat the cells with this iacs inhibitor what are the levels of these 80 different metabolites and from here we could see that there was an accumulation of reducing equivalence in nadh there was a reduction in these nucleotide triphosphates and an increase in the nucleotide monophosphates and then there was also this peculiar um decrease in aspartate that was uh pretty interesting and that they wanted to follow up on so in this particular study the focus was on three different isotopically labeled tracers glucose glutamine and aspartate so for the glucose analysis um what we can see here is that after uh in cells that are in the presence of iacs the labeled glucose is heavily funneled into lactate and alanine so there's a huge flow increased flow of glucose through the glycolysis pathway but also a reduction of the flow of glucose through the tca cycle and all these tca cycle intermediates if we take a look at the glutamine labeled case what we see here is that there's actually glucose isn't really flowing through the tca cycle glutamine is able to actually continue to flow through the tca cycle when the cells are treated with this iacs compound and then to get to the really interesting bit back to this decrease in aspartate after a treatment with iacs what we find um is that aspartate can be used uh to make some pyrimidines as i mentioned on an earlier slide and what we see here is that when we add aspartate to the cells there's actually an increase in labeling um under the condition of iacs of atp sorry i know it's hard to see down here and then that increased labeling is also found in cytodean triphosphate as well and in an aspartate rescue experiment the researchers were able to see that um iacs usually reduces cell number again by causing cell death and reducing cell proliferation but if we add aspartate to those cells there's actually um a huge regain in this cell number so there the aspartate is rescuing those cells and this study can also be done in an in vivo case so in particular uh this some acute myeloid leukemia cells were injected into mice um and then the mice were treated with vehicle or different levels of the iacs compound and cells could be taken back out of these mice and analyzed and cells analyzed from these mice that had the iacs treatment again had this lower oxygen consumption rate from the iacs treatment um so this was really cool to see in vivo um that this would also happen and then again with the iscs treatment we could look at the aspartate levels um again from samples collected from these animals and in the animals treated with higher doses of iacs there was a reduction in the levels of aspartate in the whole animal so this is really having an impact on the whole organism now and then again with uh looking at survival time in these animals we've they've the researchers found that the iacs treatment uh does increase survival time the highest doses about uh doubling survival time so that was exciting for them to see so again the seahorse measurements and lcms qualitative flux analysis was able to really zero in on the mechanism of action of this iscs compound and again glycolysis was really funneled into lactate and alanine in in these cells and glutamine was used to fuel the tca cycle where there's also this reduction in aspartate and nucleotides so essentially the iscs compound is leading to this depletion of nucleotides which as cells in encounter dna damage there's less ability to recover from this damage and less dna biosynthesis and this causes the reduced proliferation and cell death so with that um i'll summarize uh the you know the examples that i've gone over in this talk and especially want to highlight the fact that these metabolic flux measurements um really provide insights into the metabolic shifts across different cell states and i showed a bunch of different research applications in this talk but there are also a lot of other research applications that can be used for these technologies um so we can monitor t cell activation particularly in in infectious disease and cancer immunotherapy we could also monitor stem cell differentiation um there's a a huge promise for using these techniques to look at functional uh genomic screens of different crispr libraries um so there are a lot of different applications that this could be useful for and so with that um i guess i i really want to reiterate this point that um doing this type of flux analysis and really getting at the metabolic rates going on inside these cells helps us reveal what cells are doing we're not just looking at what they are but what they are actually doing and i'll leave with this last message that metabolic shifts in any living system are really causally linked to alterations in the flux through one or more different metabolic pathways so with that i'm i'm happy that you joined the seminar and i'm excited to take your questions thank you genevieve for a wonderful talk um we have a couple of questions the very uh first is uh asking about uh the sample preparation i believe so the specific question is asking uh are um are the analysis and measurements made using samples of blood samples from air or in water great yeah no i i guess i i'm sorry i didn't clarify this earlier that's a really good question so in all of the examples i show what we're doing um in the seahorse analysis is you're monitoring live cells that have been plated in uh for instance a 96 well plate um and so those are live cells sitting in culture media and you're monitoring um the glycolysis and respiration from those cells and then in the lcms uh qualitative data flux analysis um there are a lot of versions of how you can do this um but the the examples i gave in this talk were for cell culture so you're adding that isotopically labeled compound to cells in culture um and then you you remove the media after some period of time from those cells and you go through a typical lcms cellular sample prep typically this is addition of an organic solvent often done at cold temperatures to really quench metabolism quickly and then you can extract the metabolites from those cells and run those on the lcms and then in the there was that one example of the animal studies you can do these types of studies in animals as well both taking cells from those animals to put them in the sea horse or you can actually add isotopically labeled compounds to the animals you know give them an injection for instance and then take cells or samples from those animals and see what happened to those isotopically labeled compounds over time wonderful the next question is asking regarding the assays that test the metabolism of cancerous cells assuming one can inhibit the pathways that feel the oncogenes one can possibly trigger apoptosis of apoptosis cell death are you looking along those lines um yeah i think that's something of that's a of a lot of interest in the field um i think i can't say that we're specifically looking at that in our internal research programs but i think that um the technologies that i shared with you today researchers could use to understand that topic better great and the next question is asking how few cells can be used for each of these analyses seahorse and stable label isotope tracing great question um so for the seahorse analysis typically it varies uh depending on the cell type you're using and how metabolically active it is so the more active it is the easier it is to get a signal from it um typically what we're using in those seahorse assays is enough the number of cells that would roughly give a monolayer on a 96 well plate um our sea horse plates actually have a different geometry from normal 96 well plates so they're a little bit um they have a smaller the wells have a smaller diameter than typical plates um not by too much by a little bit but however many cells that would normally fit in a mono layer is as the number of cells we use for those assays um and then for the lcms analysis a common number of cells that would be used is something like a million cells per sample although i i would say you could probably go down a little bit for these particular analyses um to maybe the low hundreds of thousands of cells but somewhere around there's the limit great and do you wash cells by warm pbs or cold ah okay so we have some metabolomics sample prep people in in the audience um so that's a it's another really good question um there is a lot of debate in the field even of whether you should do washing at all and so i think that that has to be a lab specific uh determination of what they like best but yeah you could either choose not to wash the cells at all in that case you can have some leftover layer left behind so it might not be best in the isotopically labeled uh cell case um and then yes you could you could wash the cells with uh warm or cool pbs um and i there's really a lot of literature on both of those options so i would maybe internally test both of them and see if there's a major difference in particular samples and then decide how to move forward wonderful in the study where cells were cultured at 31 degrees was the seahorse still operated at 37. oh yeah that was a great question so i had to gloss over that a little bit in that particular study it was done both ways um so the cells were cultured at 37 and 31 and then the sea horse assays were red at both 31 and 37. it does
take a while for the seahorse to equilibrate to that temperature so they had to be done on on different cells but the example that i kind of hovered over with my mouse that was an example where the sea horse was run at 31 and can you also perform cell culture headspace analysis to measure gas phase and metabolites yeah i'm i'm not as familiar with that um but it should be doable with gcms technologies you should be able to do that sort of analysis i'm not sure exactly how sensitive it is or how many cells are needed for that type of analysis i'm just not as familiar with it more question how do you normalize your data for lcms analysis by protein concentration cell number or dna uh that's that's uh sorry so you said protein what was oh cell number was the second one uh okay so definitely all our options and i think it's it most likely will come down to your specific example um so a lot of people are i think are fans of the dna normalization um on the other hand when it comes to cancer models and things like that um that might be a case where your different samples are going to vary in the amount of dna the cells have um and so in that case you might want to try a protein concentration or cell number i think in in a lot of the studies i've done i've you know relied heavily on on cell number analysis or knowing how many cells go into my assays um and i get pretty tight error bars on those um but i think it's gonna come down to the specific case um and and what makes most sense um there are obviously cases where your treatment of the cells might alter the proteins that are being made in those cells and so that could that could again alter your protein content um and so yeah i think it's going to come down to this specific example wonderful we're running out of time there's a comment that we will forward to genevieve on behalf of carolina levi i believe and with that thank you all for being here for the seminar genevieve thank you very much for your time for the great talk we really enjoyed it a lot and we learned a lot thank you all have a good afternoon great thank you and thanks for having me thank you you
2021-02-20 23:50


